The
April 8, 2025
“Risk of infection due to airborne virus in classroom environments lacking mechanical ventilation” published in PLoS One
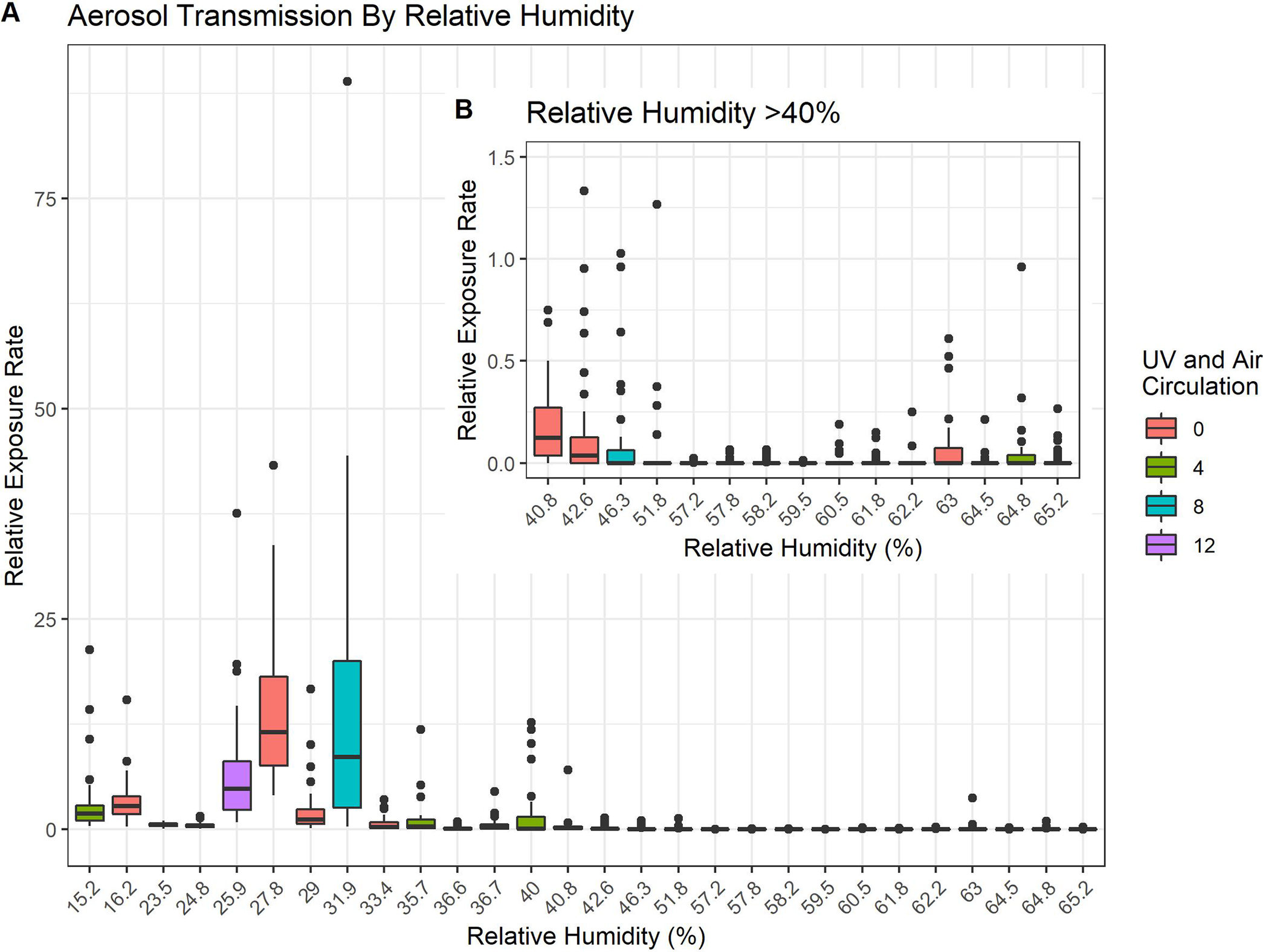
During the pandemic, Fab Spagnolo and Queens College undergraduates, Alexandra Goldblatt, Michael Loccisano and Mazharul Mahe, took advantage of the empty classrooms to test how viruses disperse in unventilated spaces. Skeptical of the CDC’s recommendations to maintain 6’ separation indoors, the team used a…